AlphaFold: Synthetic Biology and AI-Powered Protein Folding
Learning to use the Nobel prize-winning software that helped to revolutionise the way
scientists model proteins.
Why Alphafold?
The 2024 chemistry Nobel was awarded to John Jumper and Demis Hassabis at Google DeepMind
for the development of a game-changing AI tool. This AI tool, called AlphaFold, was able to predict protein structures.
Not only has AlphaFold helped reveal millions of intricate 3D protein structures, it has also helped scientists
understand how life's molecules interact, and is also completely free to start off with. I had a great time using
the software to model proteins, such as their template Protein-RNA-Ion: PDB 8AW3, but also haemoglobin and antibodies
(the genetic code for which I found on online datasets like Genbank). It’s fascinating to finally be able to see not
just the 2D representation of a structure, but the actual 3D make-up of proteins!
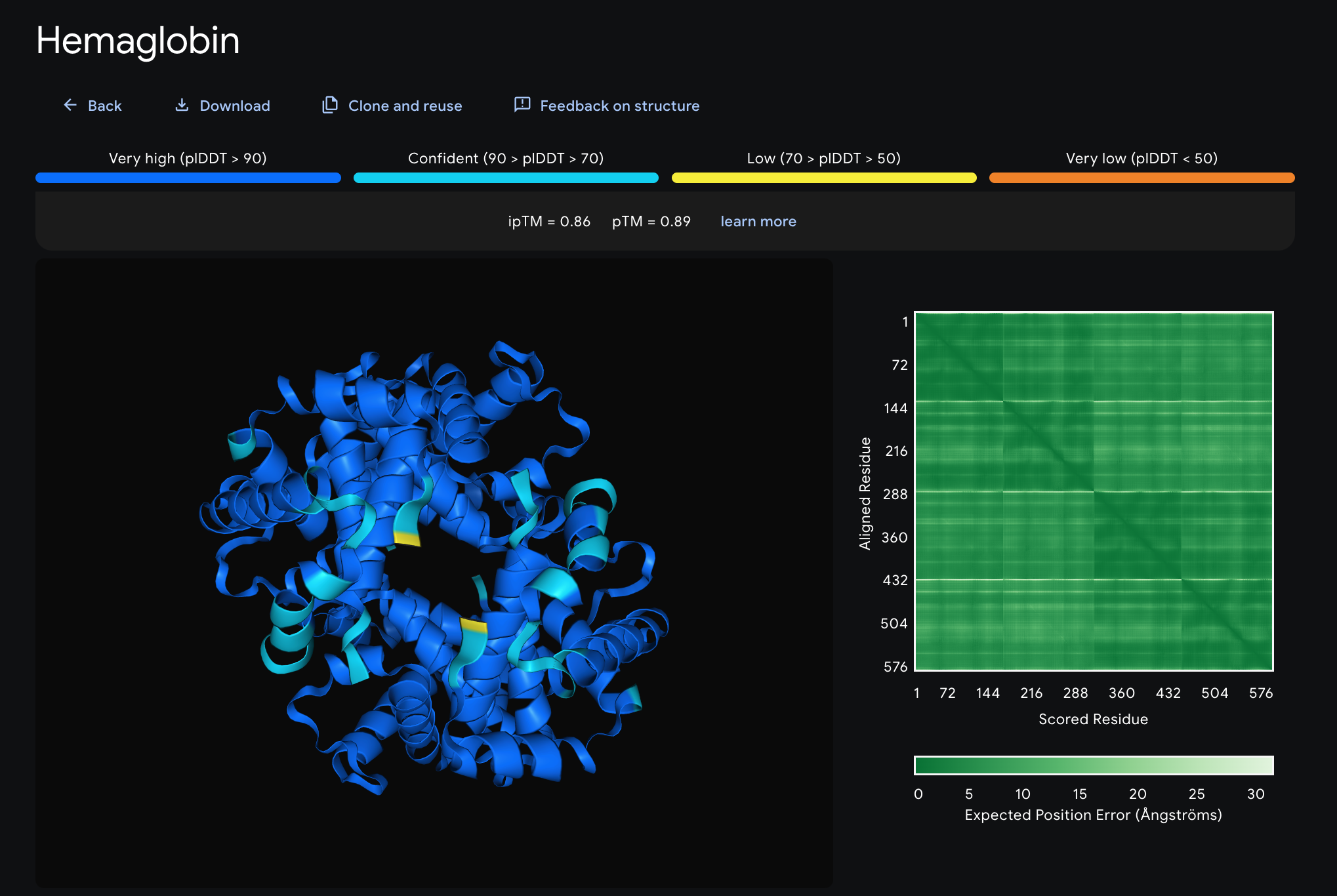
We can see that the confidence level of this is quite high, and from
what we know about haemoglobin and its shape, this algorithm has generated a pretty solid and accurate 3D visualisation
of the protein itself. According to an article, how it works is that Alphafold takes the amino acid sequence at the simplest
level and predicts the positions and bond angles of molecules in 3D space. What surprised me was that Alphafold
actually uses unsupervised learning in a way similar to GPT-3. GPT-3 learns the general features of a language
by using unlabelled text data in a similar way to how Alphafold learns embedding from the sequence of proteins
with similar functions.
Forbes has credited Alphafold as "The Most Important Achievement in AI"
and for good reason. The protein folding problem has stood as a grand challenge in the field of biology for half
a century. To determine a protein’s three-dimensional shape based solely on the one-dimensional string of molecules
that comprise it has been “one of the most important yet unsolved issues of modern science". Sure, it can theoretically
be done by hand, but the number of different configurations that a protein might fold into is astronomical.
According to Levinthal’s paradox, any given protein can theoretically adopt about 10^300 different configurations. Each
configuration could generate a different shape that would serve a different function within the body.
To give an idea of just how influential AlphaFold has been, we knew the 3D
structures for only about 17% of the roughly 20,000 proteins in the human body before the invention of AlphaFold.
Those protein structures that we did know had been painstakingly worked out by hand in a laboratory over several decades through
tedious experiments involving the likes Of X-ray crystallography and nuclear magnetic resonance, all of which require
multi-million-dollar equipment, and months or even years of brute-forced trial and error. With AlphaFold, we now have 3D
structures for virtually all (98.5%) of the human proteome. According to a Forbes article, of these, 36% are predicted
with very high accuracy and another 22% are predicted with high accuracy. With the knowledge of the shape of each protein,
scientists are now able to
see how different molecules interact with each other, which is gamechanging for fields like medicine and bioengineering!
To find out more, here are the links I was looking at to learn more about this super cool technology!
This Forbes articleAnother online article
This article in the Harvard Medicine Magazine